 Cryptogamie, Algologie
31 (4) - Pages 451-465
Cryptogamie, Algologie
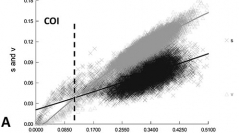
31 (4) - Pages 451-465A total of 290 florideophyte samples from the Hawaiian Rhodophyta Biodiversity Survey, spanning 17 orders of red algae, were sequenced for the mitochondrial COI DNA barcode, partial nuclear LSU rRNA gene, and plastid UPA rRNA marker. COI was A-T rich (> 60 overall) in comparison to the two rRNA markers, and was also the least conserved, with the third codon position showing the most substitution. Saturation was reached at F84 distances of approximately 0.11 for COI and 0.30 for LSU, but not at all for UPA, and some differences were found when orders or groups of families were individually examined. Rates of sequencing success for the three markers ranged from 46.8 for COI to 64.7 for UPA and 79.6 for LSU, indicating substantial differences in ease of data acquisition. Concatenation of marker sequences starting with the least saturated marker (UPA), and adding in order of degree of saturation (UPA+LSU, followed by UPA+LSU+COI) resulted in a strong increase in bootstrap support with neighbor-joining analysis, indicating that some phylogenetic utility can be gained from barcode-like sequences that are obtained for biodiversity surveys.
Also available on Connect.barcodeoflife